pacman::p_load(sf, sfdep, tmap, tidyverse, plotly, zoo, Kendall)In-class Exercise 7
Global and Local Measures of Spatial Autocorrelation - SFDEP
Overview
In this hands-on exercise, you will learn how to compute Global and Local Measure of Spatial Autocorrelation (GLSA) by using spdep package. By the end to this hands-on exercise, you will be able to:
import geospatial data using appropriate function(s) of sf package,
import csv file using appropriate function of readr package,
perform relational join using appropriate join function of dplyr package,
compute Global Spatial Autocorrelation (GSA) statistics by using appropriate functions of spdep package,
plot Moran scatterplot,
compute and plot spatial correlogram using appropriate function of spdep package.
compute Local Indicator of Spatial Association (LISA) statistics for detecting clusters and outliers by using appropriate functions spdep package;
compute Getis-Ord’s Gi-statistics for detecting hot spot or/and cold spot area by using appropriate functions of spdep package; and
to visualise the analysis output by using tmap package.
1. Installing and Loading R packages
1.1 Importing the Geospatial Data
The code chunk below uses st_read() of sf package to import Hunan shapefile into R. The imported shapefile will be simple features Object of sf.
hunan <- st_read(dsn = "Data/Geospatial",
layer = "Hunan")Reading layer `Hunan' from data source
`C:\Harith-oh\IS415-Harith\In-class_Ex\data\Geospatial' using driver `ESRI Shapefile'
Simple feature collection with 88 features and 7 fields
Geometry type: POLYGON
Dimension: XY
Bounding box: xmin: 108.7831 ymin: 24.6342 xmax: 114.2544 ymax: 30.12812
Geodetic CRS: WGS 841.2 Importing CSV file into environment
Next, we will import Hunan_2012.csv into R by using read_csv() of readr package. The output is R dataframe class.
hunan2012 <- read_csv("Data/Aspatial/Hunan_2012.csv")1.3 Performing relational join
The code chunk below will be used to update the attribute table of hunan’s SpatialPolygonsDataFrame with the attribute fields of hunan2012 dataframe. This is performed by using left_join() of dplyr package.
In order to retain the geospatial properties, the left data frame must be the sf data.frame (i.e. hunan)
hunan_GDPPC <- left_join(hunan,hunan2012)%>%
select(1:4, 7, 15)2. Plotting a Choropleth Map
tmap_mode("plot")
tm_shape(hunan_GDPPC) +
tm_fill("GDPPC",
style = "quantile",
palette = "Blues",
title = "GDPPC") +
tm_layout(main.title = "Distribution of GDP per captial by district",
main.title.position = "center",
main.title.size = 1.2,
legend.height = 0.45,
legend.width = 0.35,
frame = TRUE) +
tm_borders(alpha = 0.5) +
tm_compass(type="8star", size = 2) +
tm_scale_bar() +
tm_grid(alpha = 0.2)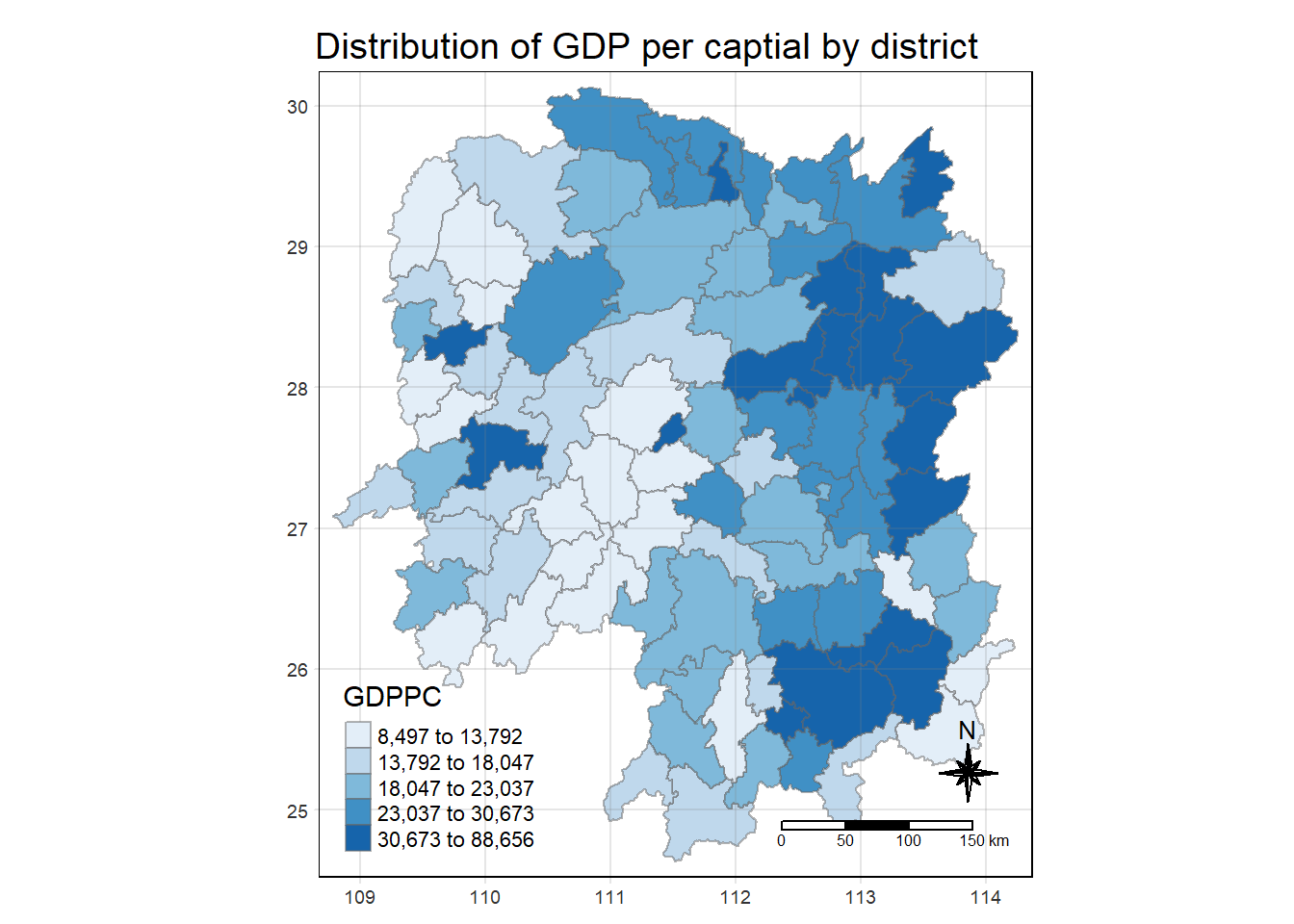
3. Deriving the contiguity weights
3.1 Contiguity weights: Queen’s method
wm_q <- hunan_GDPPC %>%
mutate(nb = st_contiguity(geometry),
wt = st_weights(nb,
style = "W"),
.before = 1)3.2 Contiguity weights: Rook’s method
wm_r <- hunan_GDPPC %>%
mutate(nb = st_contiguity(geometry),
queen = FALSE,
wt = st_weights(nb,
style = "W"),
.before = 1)3.3 Computing Global Moran I
moranI <- global_moran(wm_q$GDPPC,
wm_q$nb,
wm_q$wt)3.4 Performing Global Moran I’s Test
global_moran_test(wm_q$GDPPC,
wm_q$nb,
wm_q$wt)
Moran I test under randomisation
data: x
weights: listw
Moran I statistic standard deviate = 4.7351, p-value = 1.095e-06
alternative hypothesis: greater
sample estimates:
Moran I statistic Expectation Variance
0.300749970 -0.011494253 0.004348351 3.5 Performing Global Moran’s I permutation test
To ensure results stay the same when rendering every time
set.seed(1234)global_moran_perm(wm_q$GDPPC,
wm_q$nb,
wm_q$wt,
nsim = 99)
Monte-Carlo simulation of Moran I
data: x
weights: listw
number of simulations + 1: 100
statistic = 0.30075, observed rank = 100, p-value < 2.2e-16
alternative hypothesis: two.sidedIf observation values are small, it is better to use a higher number of simulations
3.6 Computing local Moran’s I
lisa <- wm_q %>%
mutate(local_moran = local_moran(
GDPPC, nb, wt, nsim = 99),
.before = 1) %>%
unnest(local_moran)
lisaSimple feature collection with 88 features and 20 fields
Geometry type: POLYGON
Dimension: XY
Bounding box: xmin: 108.7831 ymin: 24.6342 xmax: 114.2544 ymax: 30.12812
Geodetic CRS: WGS 84
# A tibble: 88 × 21
ii eii var_ii z_ii p_ii p_ii_…¹ p_fol…² skewn…³ kurtosis
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 -0.00147 0.00177 4.18e-4 -0.158 0.874 0.82 0.41 -0.812 0.652
2 0.0259 0.00641 1.05e-2 0.190 0.849 0.96 0.48 -1.09 1.89
3 -0.0120 -0.0374 1.02e-1 0.0796 0.937 0.76 0.38 0.824 0.0461
4 0.00102 -0.0000349 4.37e-6 0.506 0.613 0.64 0.32 1.04 1.61
5 0.0148 -0.00340 1.65e-3 0.449 0.654 0.5 0.25 1.64 3.96
6 -0.0388 -0.00339 5.45e-3 -0.480 0.631 0.82 0.41 0.614 -0.264
7 3.37 -0.198 1.41e+0 3.00 0.00266 0.08 0.04 1.46 2.74
8 1.56 -0.265 8.04e-1 2.04 0.0417 0.08 0.04 0.459 -0.519
9 4.42 0.0450 1.79e+0 3.27 0.00108 0.02 0.01 0.746 -0.00582
10 -0.399 -0.0505 8.59e-2 -1.19 0.234 0.28 0.14 -0.685 0.134
# … with 78 more rows, 12 more variables: mean <fct>, median <fct>,
# pysal <fct>, nb <nb>, wt <list>, NAME_2 <chr>, ID_3 <int>, NAME_3 <chr>,
# ENGTYPE_3 <chr>, County <chr>, GDPPC <dbl>, geometry <POLYGON [°]>, and
# abbreviated variable names ¹p_ii_sim, ²p_folded_sim, ³skewnessVariable mean and pysal value should be the same. For take home exercise 2, stay with mean
3.7 Visualising local Moran’s I
3.7.1 Computing ii
tmap_mode("plot")
tm_shape(lisa) +
tm_fill("ii") +
tm_borders(alpha = 0.5) +
tm_view(set.zoom.limits = c(6,8))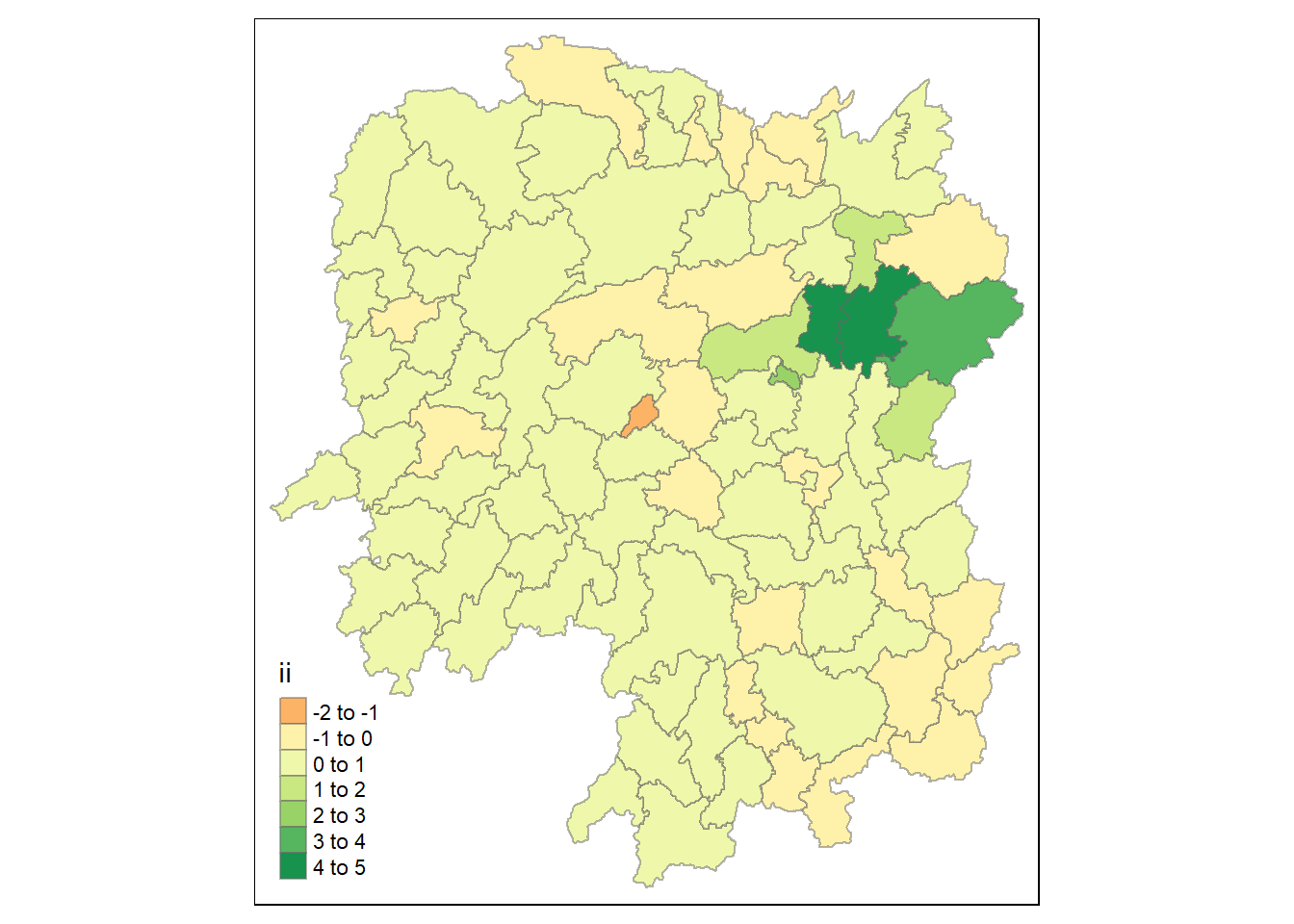
3.7.2 Computing p_ii
tmap_mode("plot")
tm_shape(lisa) +
tm_fill("p_ii") +
tm_borders(alpha = 0.5)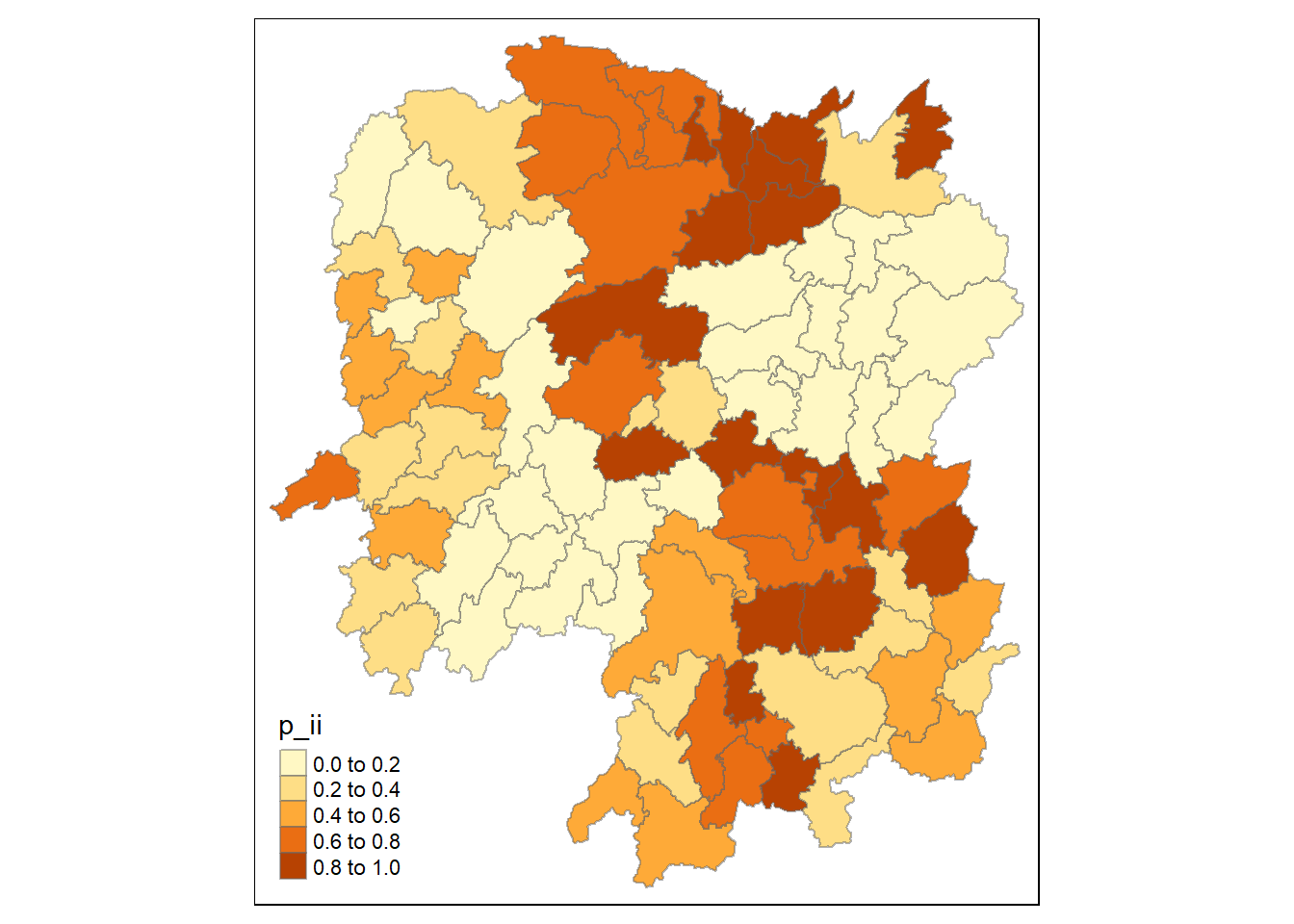
Ideally should use p_ii_sim variable of lisa so that results produced is stable.
3.7.3 Visualising the local Moran’s I Map
lisa_sig <- lisa %>%
filter(p_ii < 0.05)
tmap_mode("plot")
tm_shape(lisa) +
tm_polygons() +
tm_borders(alpha = 0.5) +
tm_shape(lisa_sig) +
tm_fill("mean") +
tm_borders(alpha = 0.4)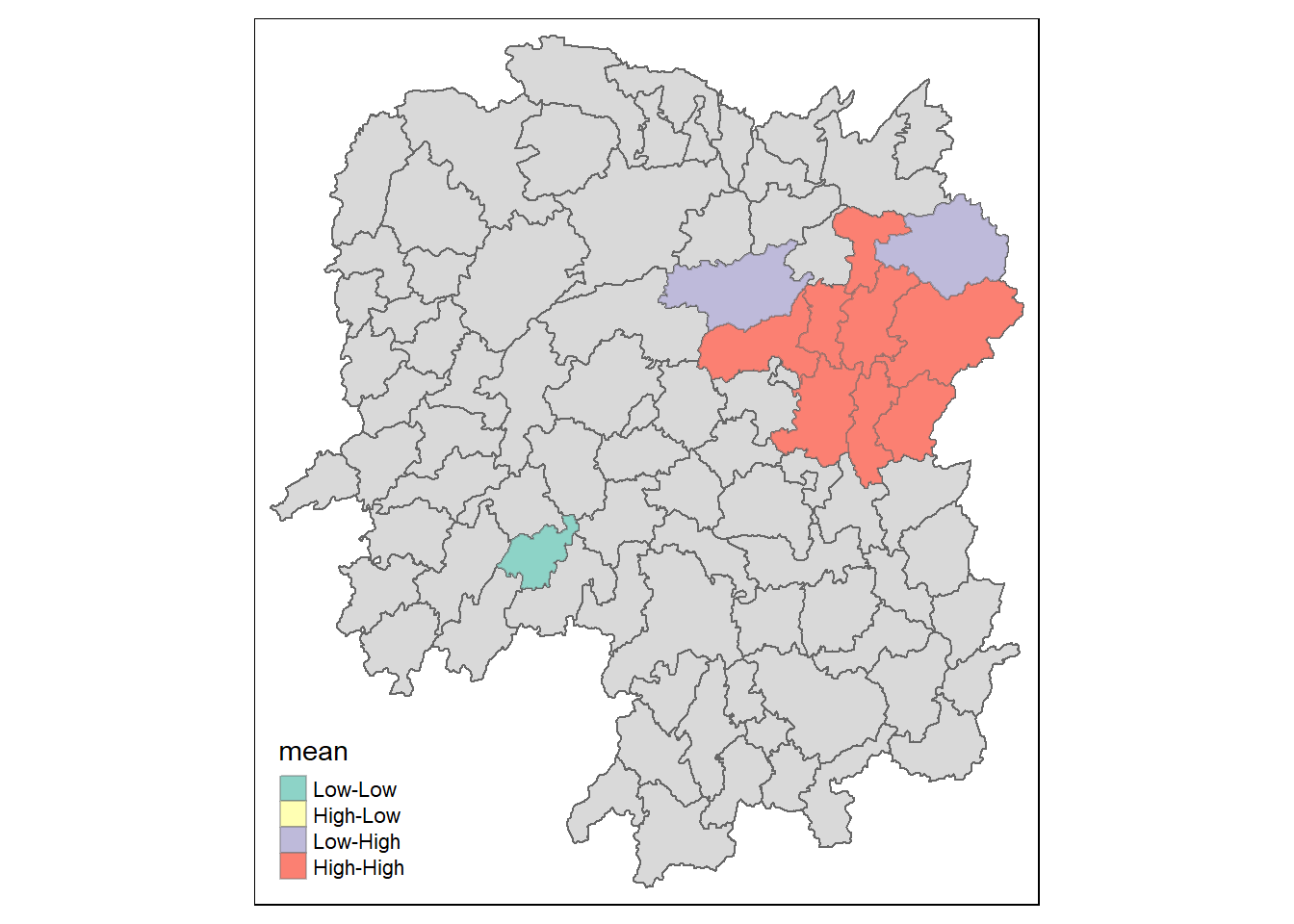
For take home exercise 2, add on to use insignificant on top of LL,HL,LH,HH, no need to use LISA but hot & cold spot areas
4. Hot Spot and Cold Spot Analysis
HCSA <- wm_q %>%
mutate(local_GI = local_gstar_perm(
GDPPC, nb, wt, nsim = 99),
.before = 1) %>%
unnest(local_GI)
HCSASimple feature collection with 88 features and 16 fields
Geometry type: POLYGON
Dimension: XY
Bounding box: xmin: 108.7831 ymin: 24.6342 xmax: 114.2544 ymax: 30.12812
Geodetic CRS: WGS 84
# A tibble: 88 × 17
gi_star e_gi var_gi p_value p_sim p_fol…¹ skewn…² kurto…³ nb wt
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <nb> <lis>
1 -0.00567 0.0115 0.00000812 9.95e-1 0.82 0.41 1.03 1.23 <int> <dbl>
2 -0.235 0.0110 0.00000581 8.14e-1 1 0.5 0.912 1.05 <int> <dbl>
3 0.298 0.0114 0.00000776 7.65e-1 0.7 0.35 0.455 -0.732 <int> <dbl>
4 0.145 0.0121 0.0000111 8.84e-1 0.64 0.32 0.900 0.726 <int> <dbl>
5 0.356 0.0113 0.0000119 7.21e-1 0.64 0.32 1.08 1.31 <int> <dbl>
6 -0.480 0.0116 0.00000706 6.31e-1 0.82 0.41 0.364 -0.676 <int> <dbl>
7 3.66 0.0116 0.00000825 2.47e-4 0.02 0.01 0.909 0.664 <int> <dbl>
8 2.14 0.0116 0.00000714 3.26e-2 0.16 0.08 1.13 1.48 <int> <dbl>
9 4.55 0.0113 0.00000656 5.28e-6 0.02 0.01 1.36 4.14 <int> <dbl>
10 1.61 0.0109 0.00000341 1.08e-1 0.18 0.09 0.269 -0.396 <int> <dbl>
# … with 78 more rows, 7 more variables: NAME_2 <chr>, ID_3 <int>,
# NAME_3 <chr>, ENGTYPE_3 <chr>, County <chr>, GDPPC <dbl>,
# geometry <POLYGON [°]>, and abbreviated variable names ¹p_folded_sim,
# ²skewness, ³kurtosis4.1 Visualising Gi*
tmap_mode("view")
tm_shape(HCSA) +
tm_fill("gi_star") +
tm_borders(alpha = 0.5) +
tm_view(set.zoom.limits = c(6,8))4.2 Visualising the p value of HCSA
tmap_mode("plot")
tm_shape(HCSA) +
tm_fill("p_sim") +
tm_borders(alpha = 0.5)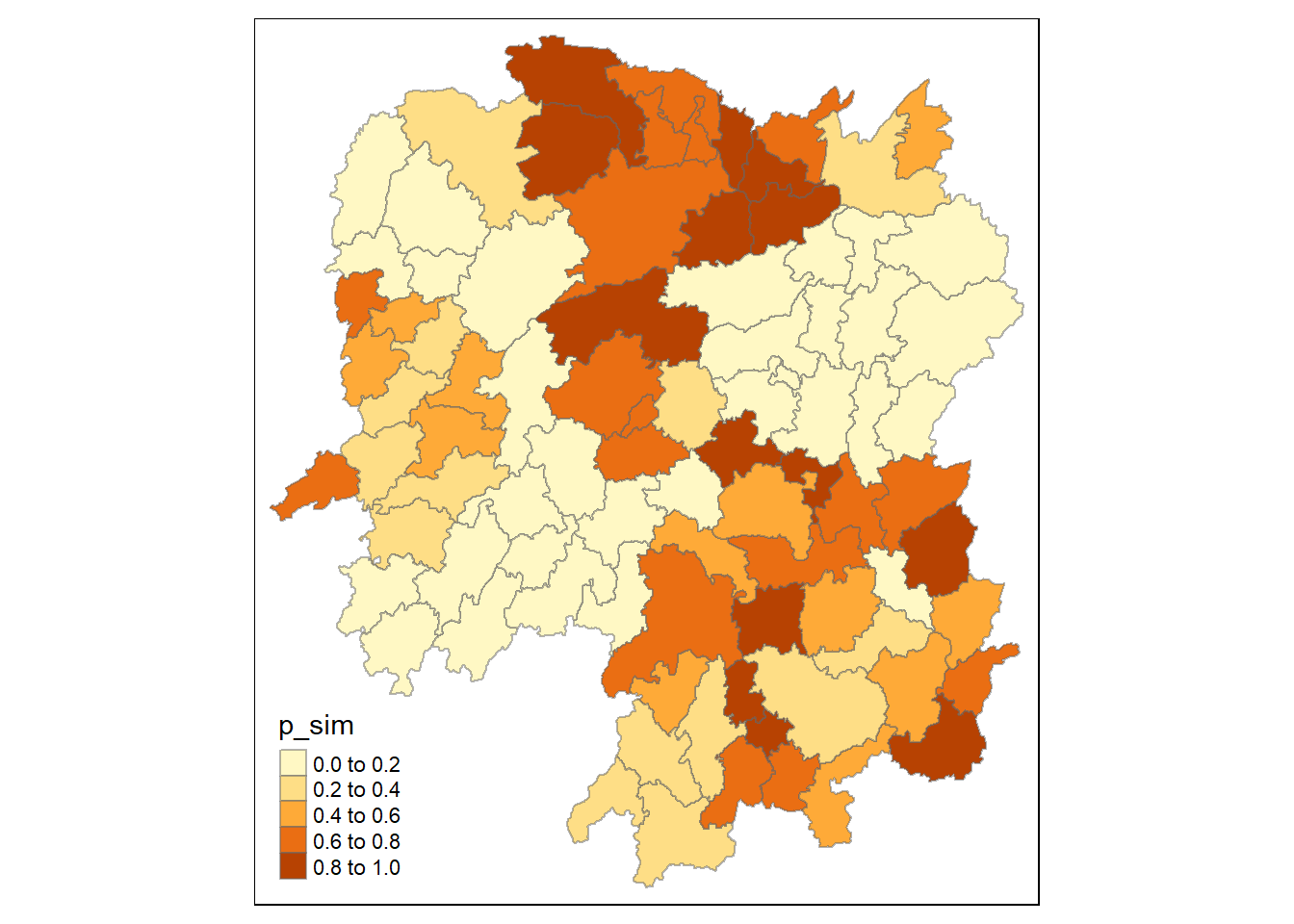
5. Mann-Kendall Test
5.1 Import files of Hunan GDPPC
GDPPC <- read_csv("Data/Aspatial/Hunan_GDPPC.csv")5.1.1 Creating a time series cube
GDPPC_st <- spacetime(GDPPC, hunan,
.loc_col = "County",
.time_col = "Year")To construct spacetime cube, we must obtain the location and time
GDPPC_nb <- GDPPC_st %>%
activate("geometry") %>%
mutate(
nb = include_self(st_contiguity(geometry)),
wt = st_weights(nb)
) %>%
set_nbs("nb") %>%
set_wts("wt")5.2 Arrange to show significant hotspot and coldspot areas
5.3 Performing Emerging Hotspot Analysis
ehsa <- emerging_hotspot_analysis(
x = GDPPC_st,
.var = "GDPPC",
k = 1,
nsim = 99
)